How to extract and plot thermodynamic properties from a GROMACS simulation
The gmx energy command
GROMACS is one of the most widely packages to perform Molecular Dynamics (MD) simulations.
In previous articles, I showed you the basic commands required to carry out the different steps of an MD simulation.
However, running a simulation should just be the tip of the iceberg. Hopefully, one would also like to extract some information from the run. Right?
One of the main strengths of GROMACS is that it also comes with a broad range of tools to perform analysis on your system.
In this article, I will show you how to extract the thermodynamic properties from your simulation using the gmx energy command.
The gmx energy command
During a simulation, it is important to monitor a series of different thermodynamic properties to always make sure that everything is proceeding according to our plans. To this aim, we use the gmx energy command.
This command simply takes an edr file as input and returns an output file in the xvg format which can be used to plot our data. Later I will show you how to plot them using Python or Xmgrace.
The syntax is quite easy:
|
|
- Call the program with
gmx - Select the
energycommand - Select the
-fflag and provide the edr file from your simulation (simulation.edr) - Call the
-oflag and decide how you want to name the output file (property.xvg)
Once you launched the command you will be asked to choose which property you want to obtain. Something like this code block below.
|
|
From here, you can select the property you want to extract by simply typing the corresponding number followed by a 0.
To obtain an xvg file for the temperature you can type:
|
|
Plotting data in the xvg format
Now that we have our xvg file we can create a plot to observe how the selected property evolved along the simulation.
This is particularly important during the equilibration phase of our experiment. During this step, we have to monitor different properties of our system to make sure that everything is correct.
Here I will show you two different ways to plot an xvg file format.
Xmgrace
The first tool we are going to use to plot the data in the xvg file is Grace.
This plotting program is perfectly able to handle a file in the xvg format. It is quite fast and intuitive and it is particularly useful when you just need a quick plot.
You can install it on Mac with homebrew.
|
|
On Linux you should be able to download it with this command:
|
|
Let’s assume that you generated a volume.xvg file that you want to investigate, and that you have Grace installed. You can easily plot the file with the following command:
|
|
As a result you will get a plot like the one displayed below.
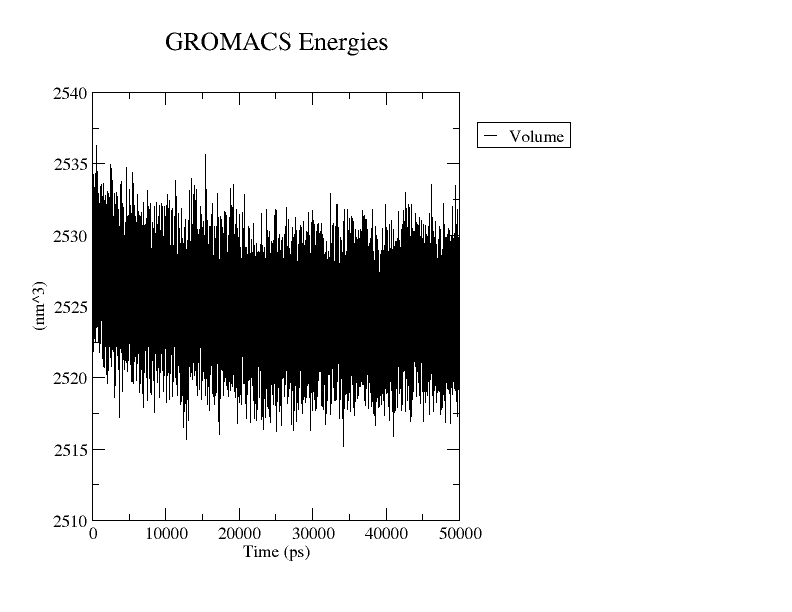
Volume variation as a function of time ($ps$) plotted with Grace
To save the image in the .png format you can enter the following:
|
|
Python
if you are more of a Python type don’t worry. You can still plot an .xvg file using numpy, and matplotlib.
|
|
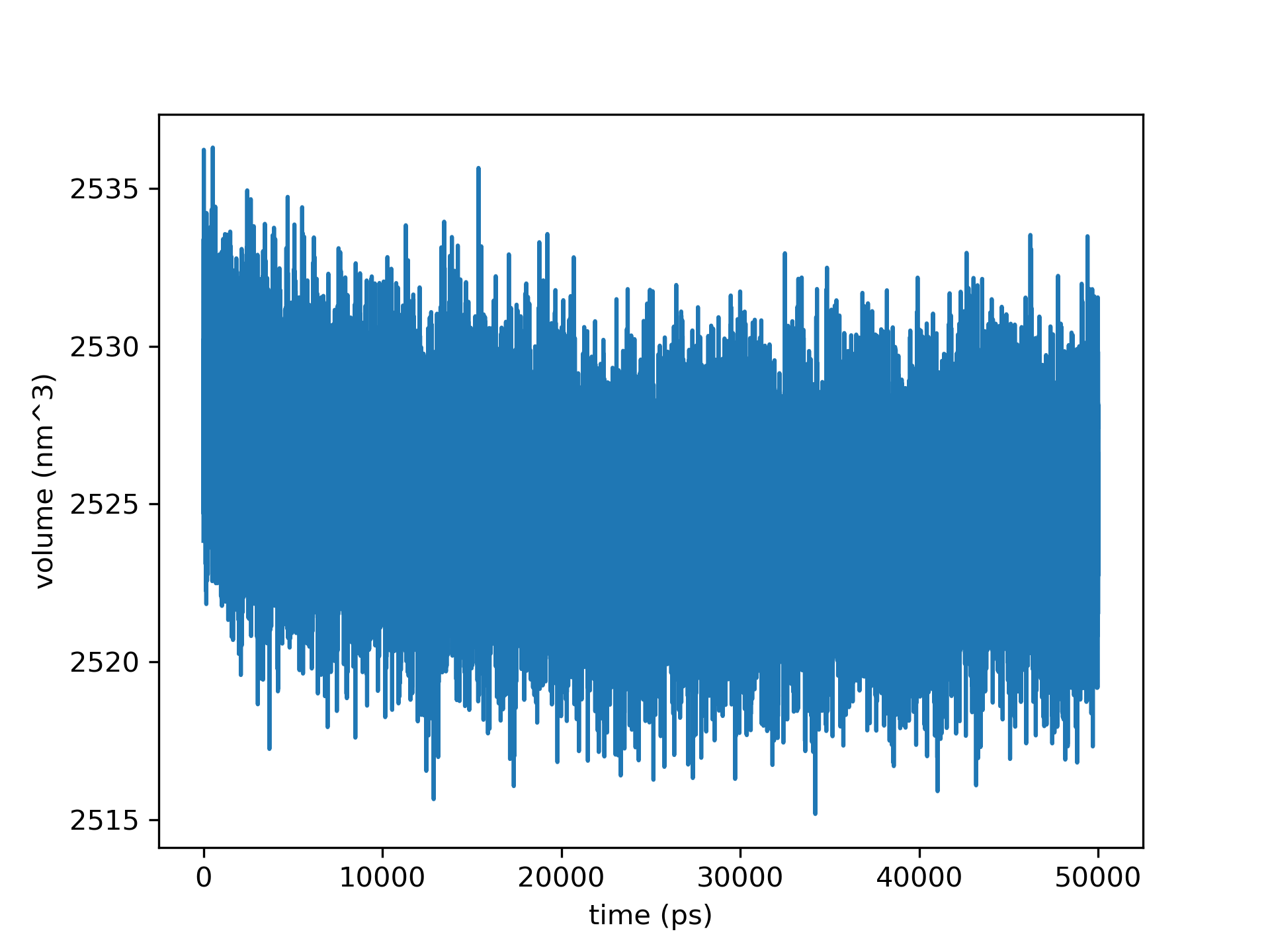
Volume variation as a function of time ($ps$) plotted with Python
Example
Here I will show you a realistic example of how to retrieve information using the gmx energy command, and the Grace plotting tool.
Let’s say that you have just finished your equilibration step in the NPT ensemble, and you want to check that everything looks right.
The first step is to take the generated edr file (that we will call npt.edr) and extract relevant properties. A common practice is to consider at least six quantities:
- Temperature
- Volume
- Pressure
- Kinetic energy
- Potential energy
- Total energy
So for each one of them you need to generate the corresponding .xvg file. You can do this by typing the following six commands.
|
|
For each line, you will be asked to select the property followed by a 0. In the end, you will have six xvg files.
You can generate a png image with the corresponding plot for each of the files with a simple bash script.
Create a text file (images.sh) and paste the following code.
|
|
Run it with bash
|
|
Now you have your images containing the plots to monitor the quantities of your simulation.
Another common property to monitor during an equilibration step is a structural feature named Root Mean Square Deviation (RMSD). Here I show you how to compute the RMSD using GROMACS.
How to merge two edr energy files
In some cases, you may happen to run a simulation for some time without getting relevant results. In such circumstances you can decide to extend the simulation for some additional time.
When extending the simulation you can result in two different energy edr files.
We can use the gmx eneconv command to merge them. The resulting edr file can be used to extract the properties from the entire simulation.
|
|
- Call the program with
gmx - Select the
eneconvcommand - Select the
-fflag and provide the two edr file from the simulations that you want to join (simulation_1.edr simulation_2.edr) - Call the
-oflag and decide how you want to name the output edr file (merged.edr)
- I generally prefer to interactively specify the starting time for each file manually via the
-settimeflag.